[Sorry for the delay in sending these solutions, but I managed to lose my phone on the train earlier today. Fortunately, someone found it, and I got it after teaching this morning’s class, but it definitely ate into my schedule…]
Back in the mid 1990s, when I worked at Time Warner’s “Pathfinder” Web site, I was asked to review some icons for weather reports that we were putting out. I saw an icon for snow, another for rain, another for fog, and another for … dust? I hadn’t ever seen a forecast of “dust” before. People from other areas of the US told me that yes, sometimes you get dust storms, and they turn the sky a weird color.
Sure enough, after moving to Israel, I experienced such dust storms. And they are weird, let me tell you. They really stick in your mind.
So when I started to see lots of orange-tinted pictures on social media from friends in New York, I wondered whether dust storms had somehow made it to Manhattan. But no, it was much worse: Large wildfires in Canada were producing such huge quantities of smoke that oodles of people across the northern United States were being told that the air outside was unsafe, and that they should wear masks if they’re outdoors.
Only in China had I previously seen people wearing masks outdoors. On one trip, when the pollution was particularly bad, I decided that I didn’t need a mask, since I would be there for a very short time. That was dumb; I returned home with a cough that lasted for several weeks.
I thus thought that it would be interesting to look at the air quality in three northern states over the last few months, and see if we could spot any obvious trends.
Data and questions
As I wrote yesterday, this week's data comes from the US Environmental Protection Agency. I downloaded it from this page:
https://www.epa.gov/outdoor-air-quality-data/download-daily-dataWe’ll be looking at PM2.5 pollution in New York, Pennsylvania, and Ohio, at all sites in all of 2023. You’ll need to download all three CSV files; I’m going to assume that you’ll name them XX_data.csv, where XX is the two-letter state name for the state data you want.
I gave you 11 tasks and questions this week. Here are my answers, along with a link to the Jupyter notebook containing my solutions:
Create a single Pandas data frame from the three downloaded files. We'll only need a few of the columns: Date, PM2.5 concentration, site name, state, longitude, and latitude. Make the index a combination of the date and state name. Rename the columns to be all lowercase and shorter, to make it easier to work with.
Given the three downloaded CSV files, how can we create a single data frame? My favorite technique continues to be a list comprehension, combined with the “glob” module in the standard library and our favorite read_csv function:
import pandas as pd
import glob
all_dfs = [pd.read_csv(one_filename,
usecols=['Date',
'Daily Mean PM2.5 Concentration',
'Site Name',
'STATE',
'SITE_LONGITUDE',
'SITE_LATITUDE'],
parse_dates=['Date'])
for one_filename in glob.glob('??_data.csv')]I started off by importing the Pandas library (using the standard “pd” alias), and then glob, as well.
I then went through each of the files with two characters, followed by “_data.csv” in the current directory. Given a filename, I then invoked pd.read_csv on it, indicating which subset of columns I wanted to import, and also that the “Date” column should be interpreted as a datetime.
The result of a list comprehension is always a list. In this case, it’s a list of data frames. That’s not normally something we think about, but here it’ll be perfect, because we can then invoke “concat” on the list of data frames, returning a single data frame:
df = pd.concat(all_dfs)
df = df.rename(columns={
'Daily Mean PM2.5 Concentration':'pm25',
'SITE_LATITUDE':'latitude',
'SITE_LONGITUDE': 'longitude',
'Site Name': 'site',
'STATE':'state',
'Date':'date'})I then invoked “rename” to rename our columns; the keyword argument “columns” let me pass a dict of the original column names and the new names. Note that if you get the original column name wrong, you won’t raise a warning or exception; your renaming will be silently ignored.
Finally, I want to set the index on this data frame to be based on the “date” and “state” columns — a two-part multi-index. We can do that with:
df = df.set_index(['date', 'state'])Our data frame is now in place, ready for us to perform some analysis.
What were the minimum, median, and maximum PM2.5 particle counts measured in these three states?
Let’s start with something simple, namely getting some basic descriptive statistics on PM2.5 counts in each of the three states.
We want to call three aggregation methods on each state. This sounds like a “groupby” problem, and indeed that’s where I went with it:
df.groupby('state')['pm25'].agg(['min', 'median', 'max'])The above code asks Pandas to calculate the min, median, and max aggregation functions for each of the states in our data frame.
There are two particular things to notice here:
First, because I want to run several aggregation methods, and not just one, I have to use the “agg” method. I then pass a list of what I want to run, either as strings or as functions.
Second, notice that we’re grouping on the “state” column… which isn’t really a normal column any more, but which is part of our index. We can’t retrieve values from the column directly any more, but we can use “groupby” on it, which is great. This is what I get:
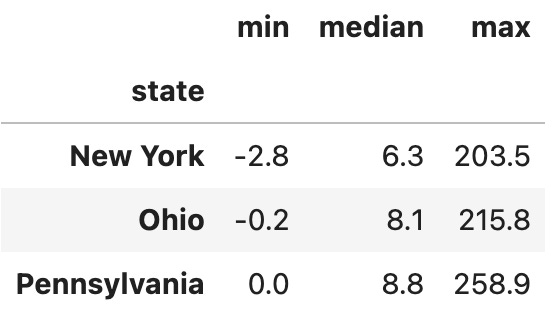
As you can see, the PM2.5 particle counts are normally quite low. (How they’re negative is beyond me, I’ll admit!) But you can see that the max values are way, way higher than what we would normally expect to see, at about 30x the median level. Something was clearly weird during this time.

