This week, in consideration of the many people (like me!) traveling to the US for PyCon, decided to examine the data produced by the International Trade Administration describing how many people have entered the US each month since the year 2000.
The data is in a single Excel spreadsheet, which you can get from here:
/content/files/sites/default/files/2022-02/monthly-arrivals-2000-present.xlsx
I asked you to start by creating two data frames from this Excel file:
- In the smaller data frame (countries_df), there are only two columns. The index contains country names, and the one non-index data column (region) comes from Excel's column B. You'll have to ignore a bunch of blank/empty rows in the Excel spreadsheet.
- In the larger data frame (travel_df), there are many columns, once for each monthly report. Make the index the same as in countries_df, with country names from Excel's column B. The other columns will all be datetime values, with a year and month. (The day doesn’t matter, and the time will always be midnight.)
Then there will be some other cleaning:
- The final columns in the file have the word “Preliminary”; that word should be stripped.
- All column names/headers should be turned into datetime objects. Remove any columns whose names that cannot be turned into datetimes.
- In the data, replace any instances of ' - ' (i.e., a string containing one space, a minus sign, and three spaces with 0.
- In the data, replace NaN with 0.
- Set the dtype of all columns to int.
I then asked you to answer the following questions:
- In the most recent report, which 10 countries had the greatest number of tourists enter the US?
- In the first report, which 10 countries had the greatest number of tourists enter the US?
- Total the number of tourists from each region in the earliest report vs. the latest report. (Yes, you could get this directly from the original Excel spreadsheet, but I want you to calculate this yourself!) Do we see any changes in the last two decades or so?
- Have any countries had more month-to-month declines in tourism to the US than increases?
- Calculate the mean of tourists from each country for each decade. (And yes, the current decade will be listed as December 31st, 2030.)
Let’s get to it!
Create countries_df: The index contains country names, and the one non-index data column (region) comes from Excel's column B. Ignore blank/empty rows.
Before starting anything else, I need to load up Pandas:
import pandas as pdI then wanted to create the smaller data frame based on the Excel spreadsheet, containing just the countries (as the index) and the regions (as the sole column). In order to do that, I can use read_excel, which can either take a filename (as a string) or a URL (also a string):
url = '/content/files/sites/default/files/2022-02/monthly-arrivals-2000-present.xlsx'
countries_df = pd.read_excel(url)At a basic level, the above code will work, creating a data frame. But this data frame contains more information than we need. The problem is that the Excel file contains many different layers and types of information, above and beyond the per-country statistics. To a person, it’s easy to read and navigate this spreadsheet. But if we want to read it into Pandas and analyze the data, we’ll need to specify exactly what we want, and from where.
For starters, since we are only interested in the countries, we can ignore the first 20 lines of the file. We can pass the “skiprows” keyword argument to read_excel, which will do exactly that.
But read_excel, like its cousin read_csv, will always try to name the columns based on the first row it does read. We don’t need those column headers, because in countries_df, we’ll only have the index (with countries) and the region. We can tell read_excel to ignore the header (with header=None), to use the columns at indexes 1 and 2 (with usercols=[1,2]), and to name the two columns (with names=['country', 'region']):
countries_df = pd.read_excel(url,
skiprows=20,
header=None,
usecols=[1,2],
names=['country', 'region'])This works fine, except that we want the “country” column to act as an index. I could run “set_index” on an existing data frame, but I can do that right away within read_excel by passing index_col:
countries_df = pd.read_excel(url,
skiprows=20,
header=None,
usecols=[1,2],
names=['country', 'region'],
index_col=0)Notice that I can pass index_col the numeric index of the column I wish to use. Also note that this number is counted from the columns actually added to the data frame, not in the original spreadsheet. Thus, while “country” is at column index 1 in Excel, it’s at column index 0 in the data frame.
Finally, I want to get rid of any row that has NaN values. We’re dealing with a very small data frame here, and it’s just to be able to associate countries with regions. Any NaN is just going to give us trouble:
countries_df = pd.read_excel(url,
skiprows=20,
header=None,
usecols=[1,2],
names=['country', 'region'],
index_col=0).dropna() This is what the first rows of this data frame look like in my Jupyter notebook:
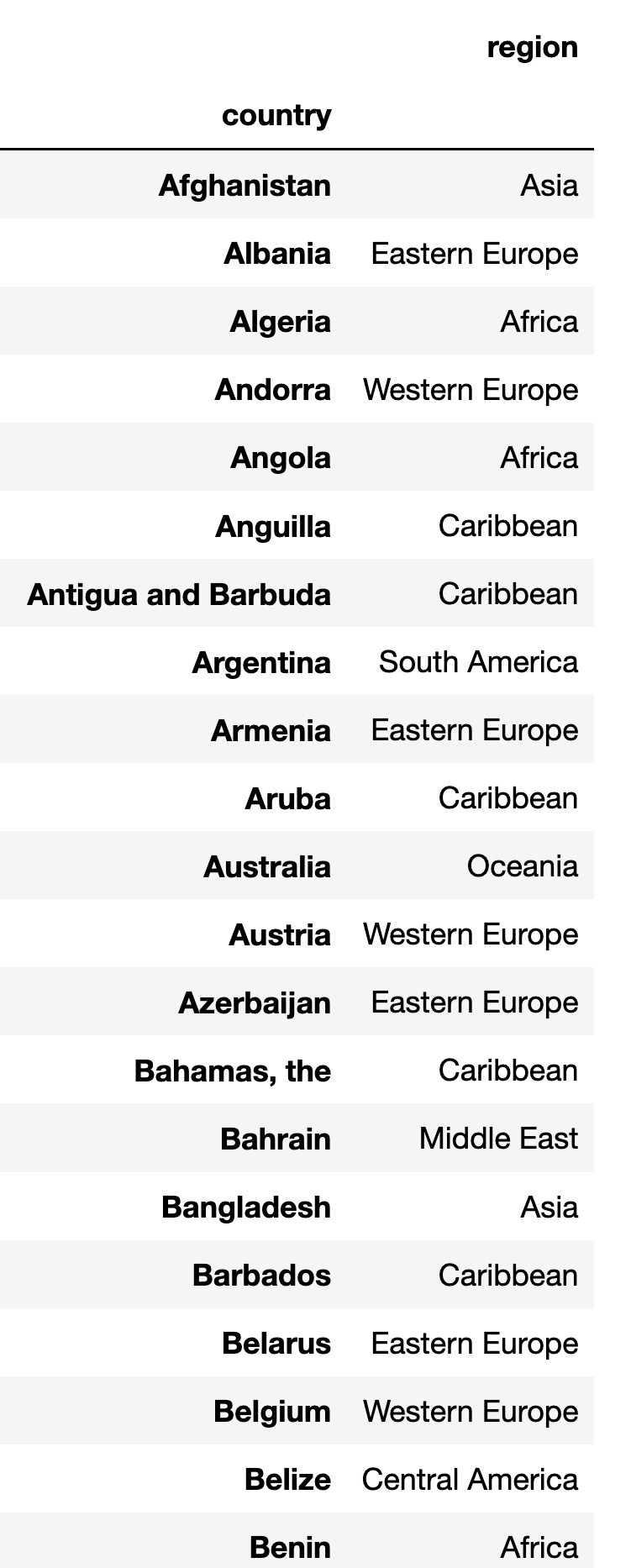
With this out of the way, we can go onto the bigger cleaning headache.
Create travel_df, with many columns, one for each monthly report. Make the index the same as in countries_df, with country names from Excel's column B. The other columns will all be datetime values, with a year and month.
There’s a lot packed into this. And really, whenever you’re dealing with data from the real world, there will be a lot of cleaning work to do. Sometimes an awful lot of it.
So let’s start by reading the Excel spreadsheet into a new data frame, and then chipping away at the problems with it:
travel_df = pd.read_excel(url)Once again, we have a bunch of rows that we don’t want, with blank space and regional information. Couldn’t I use skip_rows here, as I did with countries_df?
Not really, because skip_rows tells the parser to ignore those rows entirely when reading the Excel file into memory. The thing is, I want the first row, so that I can use it for headers. And skip_rows doesn’t really allow for that.
Instead, I’ll read the data frame into memory and drop the first 19 rows. Ironically, because the first row was used for headers, I don’t need to skip over it, and can just remove those first 19 rows:
travel_df = pd.read_excel(url).drop(range(0,19))Notice that “drop” doesn’t actually modify the data frame, but returns a new one. In this case, we invoke drop on the data frame we got back from read_excel. The result is then assigned to travel_df.
And indeed, I next get rid of four columns whose names aren’t going to be datetime values, and which don’t contain any travel information. I do this by using “df.drop”.
As we see, drop can be used to remove either rows or columns. By default, it removes rows, but if you pass “axis='columns' ” it’ll drop the named column(s) instead:
travel_df = travel_df.drop([1, ' ',
'World \nRegion', 'Notes:'], axis='columns')As you can see, the spaces and newlines in Excel have to be matched exactly in order for this to work. What a pain!
Also notice that df.drop, like so many other Pandas methods, can take either a single string (for one column) or a list of strings (for multiple columns).
Next, I want to rename the first column to be “country”. By default, it has the comically long name of
"International Visitors--
1) Country of Residence
2) 1+ nights in the USA
3) Among qualified visa types"I mean… really?
How can I rename this column? The best method is to use “rename”. We can pass it a dict in which the keys are the old column names and the values are the new column names, as in:
a_df.renane(columns={'a':'b'})Note that you have to pass “columns” as a keyword argument, since “rename” can be used in a few other ways and contexts, also.
The thing is, the old name is pretty ugly. Do I want to write it all out as a dict key? Of course not! We can retrieve its name, and then rename based on it:
travel_df = travel_df.rename(columns={travel_df.columns[0]: 'country'})I created a dict with a single key and value. The key? The first element of df.columns, which returns the columns in the data frame. The value will then be the string “country”, which is exactly what I want.
Like so many other methods in Pandas, “rename” returns a new data frame, which we then assign back to the original variable.
I then want to make this “country” column into the index. I can do that by invoking “set_index”, specifying that we want to use “country”:
travel_df = travel_df.set_index('country')This is all great, except for one little thing: Because the “country” column contained some NaN values, we now have several rows whose index is NaN, and whose values are completely worthless. These are the final three rows, and you could argue that I should just use “drop” to get rid of them.
But I decided to use a slightly more complex system, just to show you how versatile indexes are — they’re very similar to series, and can handle a lot of sophisticated operations.
In particular, I can run “isna” on the index, finding out which values are NaNs:
travel_df.index.isna()Then I can use “loc” to retrieve those rows that don’t have a NaN index.
travel_df = travel_df.loc[~travel_df.index.isna()]Notice my use of ~, the tilde, which flips the logic of every element I got back from travel_df.index.isna(). Meaning that we’ll drop the rows with NaN indexes, and keep the normal ones.
Next, I want to remove the columns whose names start with “Unnamed”. Why would we have such columns? Who knows? But they exist, and we have to get rid of them.
Remember that we can drop a list of columns by passing a list of strings to “drop”. But how can I get a list of all column names that start with “Unnamed”?
I could iterate over each of the columns, and keep track of those that start with “Unnamed”. And then I could pass that list to “drop”.
That’ll work, but I think it’s more idiomatic to use one of my favorite Python constructs, a list comprehension:
[one_name
for one_name in travel_df.columns
if one_name.startswith("Unnamed") ]If you’ve never seen a list comprehension before, then the above probably looks really weird and daunting. The basic idea is:
- We go through each element of travel_df.columns
- Does it start with “Unnamed”?
- If so, then return the name.
The result is a list of strings, column names we can use.
At least, that’s the theory. In reality, many of our column names are actually datetime values. Which means that they don’t support the “startswith” method. So we’ll get an error.
We’ll need to change our condition to turn the value into a string and then check what it starts with:
[one_name
for one_name in travel_df.columns
if str(one_name).startswith("Unnamed") ]This works just great! As always, a list comprehension creates a list. And we can pass that list to “drop” as an argument. Together with “axis='columns' ”, and assigning the result back to travel_df, we get rid of the bad columns:
travel_df = travel_df.drop([one_name
for one_name in travel_df.columns
if str(one_name).startswith("Unnamed") ],
axis='columns')